【Added on November 18, 2022】
- The Contents of this report are current as of the date of publication (December 7, 2021). Please note that these may differ from the latest trends.
- Raw data from the analysis presented in this report is now available. Please feel free to download and make use of it.
Outline
It will soon be two years since the first SARS-CoV-2 infection (COVID-19) was reported in Wuhan, China, in December 2019. COVID-19 caused a global pandemic, accompanied by the emergence of new coronavirus variants with strong infectivity. Although the pandemic in Japan is showing signs of convergence, the global spread of the virus has not yet been halted.
In the midst of the COVID-19 pandemic, development of therapeutic and prophylactic agents is proceeding at a rapid pace in Japan and abroad. One effective option is antibody drugs based on “neutralizing antibody” that has the function of preventing viral infection and multiplication. Some antibody drugs originating overseas have already been approved (Supplementary info. 1). All of these antibodies target the receptor binding domain (RBD) of SARS-CoV-2 spike protein (S protein). It is human IgG-type recombinant antibodies from antibody genes extracted from antibody-producing cells (B cells) in blood of patients who have recovered from severe acute respiratory syndrome (SARS) caused by SARS-CoV-2 or a closely related virus of SARS-CoV-2. Although some variants have been reported to escape neutralizing antibodies acquired by recovering or vaccinated individuals, antibody cocktail therapy is effective against a wide range of variants by administering two or more antibodies simultaneously.
Since last February, we have been working on the research and development of COVID-19 under the Fukushima Translational Research Project (Fukushima project). To obtain genes for antibodies against SARS-CoV-2, blood donor volunteers were recruited from patients who had recovered from COVID-19. In April this year, the Fukushima project announced that it had successfully obtained 18 neutralizing antibodies and their genes from the blood. Several groups, including Keio University, Hiroshima University, Toyama University, Osaka University, and Kumamoto University, have successively reported the acquisition of neutralizing antibodies or infection-enhancing antibodies derived from infected patients. The most significant feature of the results of the Fukushima project is the inclusion of human IgA-type antibodies in the neutralizing antibodies obtained (Supplementary info. 2). We are seeking approval of this IgA-type antibody as a pharmaceutical product. If the IgA antibody is officially approved as a drug, it will be the world’s first IgA antibody drug.
To obtain genes for antibodies against SARS-CoV-2, the Fukushima project uses a proprietary protein microarray system. The protein microarray from the Fukushima project have spotted more than 200 antigens of SARS-CoV-2. S proteins include not only full-length recombinant proteins, but also subunit-only proteins such as S1 and S2, and fragment proteins consisting only of RBD. It also includes antigens for various variants, such as alpha, beta, gamma, and delta, in addition to the conventional strains. Many antigens of coronavirus genus viruses other than SARS-CoV-2 have also been spotted. It contains antigens for SARS coronavirus and MERS (Middle East Respiratory Syndrome) coronavirus, coronaviruses that cause common colds, and coronaviruses that infect non-human animals such as a cattle and chickens. By reacting the blood of COVID-19 recovered individuals with these antigens, the same system can be used to evaluate whether antibodies against SARS-CoV-2 were produced by the infection, and if so, what antigens they react to, and whether the antibodies produced from the acquired antibody genes react to the appropriate antigen for the purpose. In addition, the protein microarray also spots about 12,000 samples, including antigens such as intestinal bacteria, allergens, and human proteins (auto-antigens). Thus, this protein microarray can be used to comprehensively examine a profile of antibodies in the blood of COVID-19 recovering individuals against a wide variety of antigens at once.
In this report, we will present results of blood antibody profiling in COVID-19 recovering patients obtained so far in the Fukushima project. Results of healthy (non-infected) specimens whose blood was drawn before the COVID-19 pandemic are also included for comparison.
About Specimen
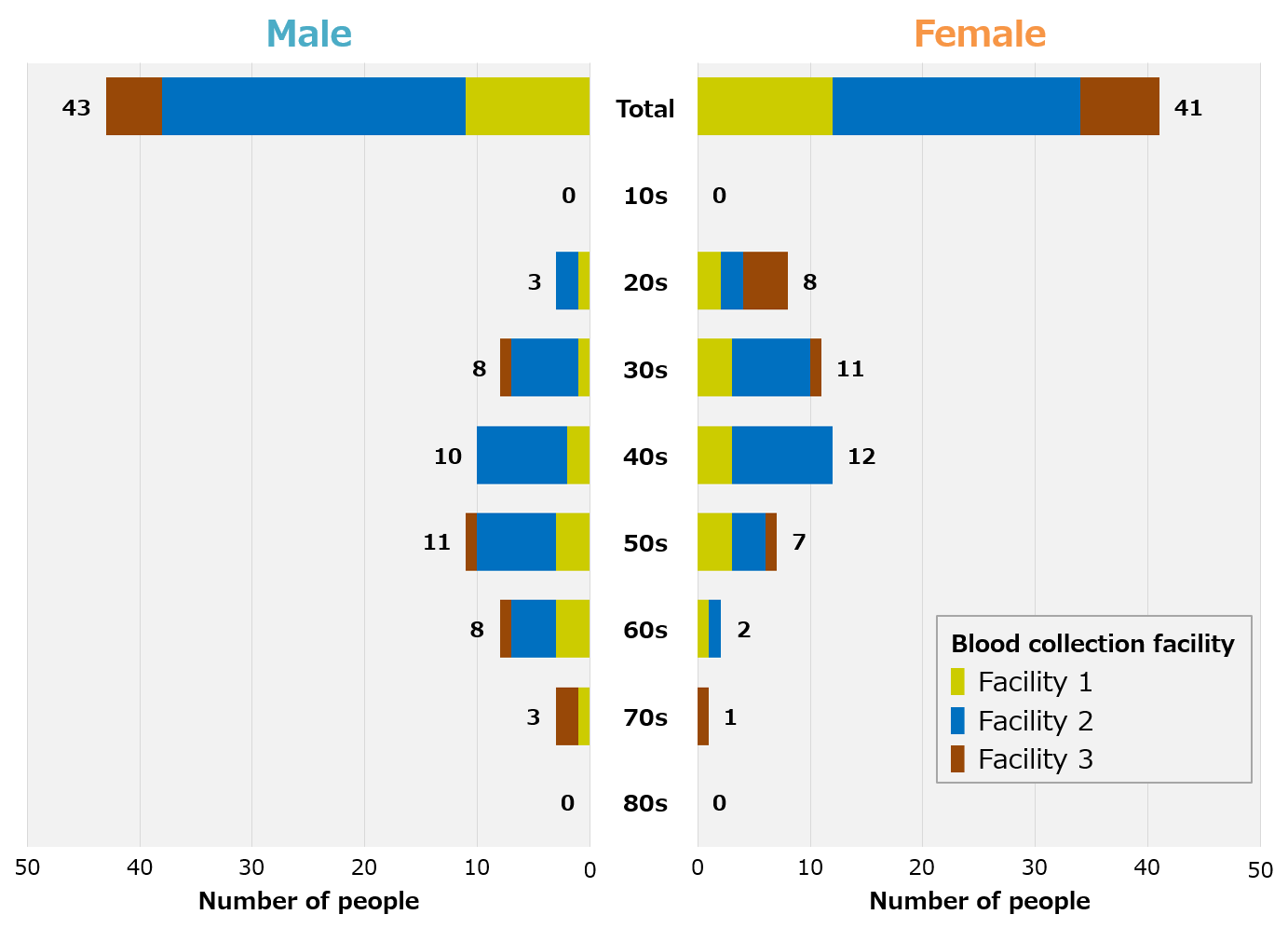
We recruited blood donor volunteers in Fukushima Prefecture and Tokyo for patients recovering from COVID-19, and were able to collect blood samples from a total of 84 people, 43 men and 41 women in their 20s to 70s (Figure 1). In addition, blood samples from healthy individuals were procured from overseas prior to the COVID-19 pandemic. Therefore, there are racial differences between recovering and healthy individuals.
■ Specimen information(Click the button to download)
| Title | File format |
| Age distribution of COVID-19 recovering individuals who donated blood. | Excel |
Results
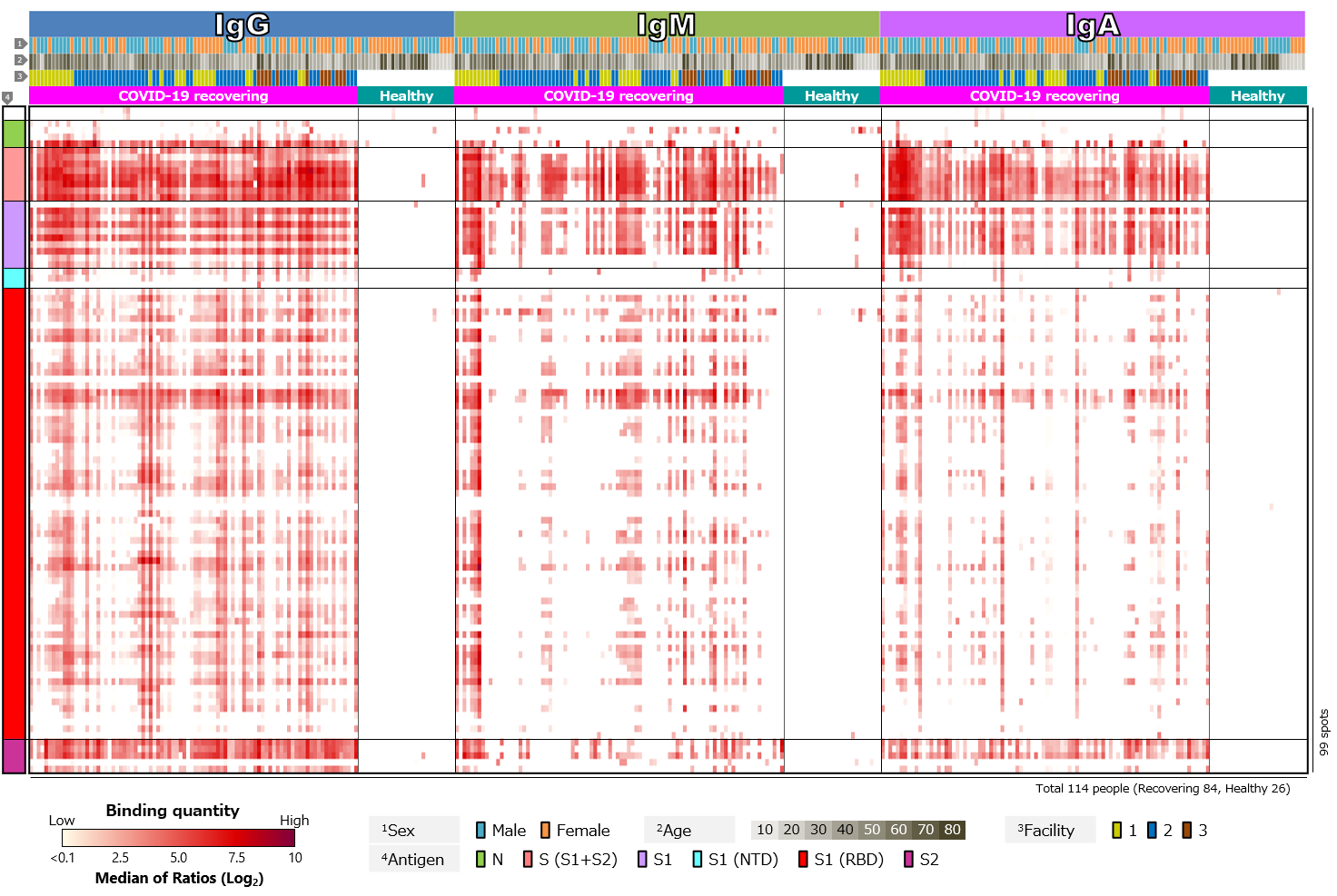
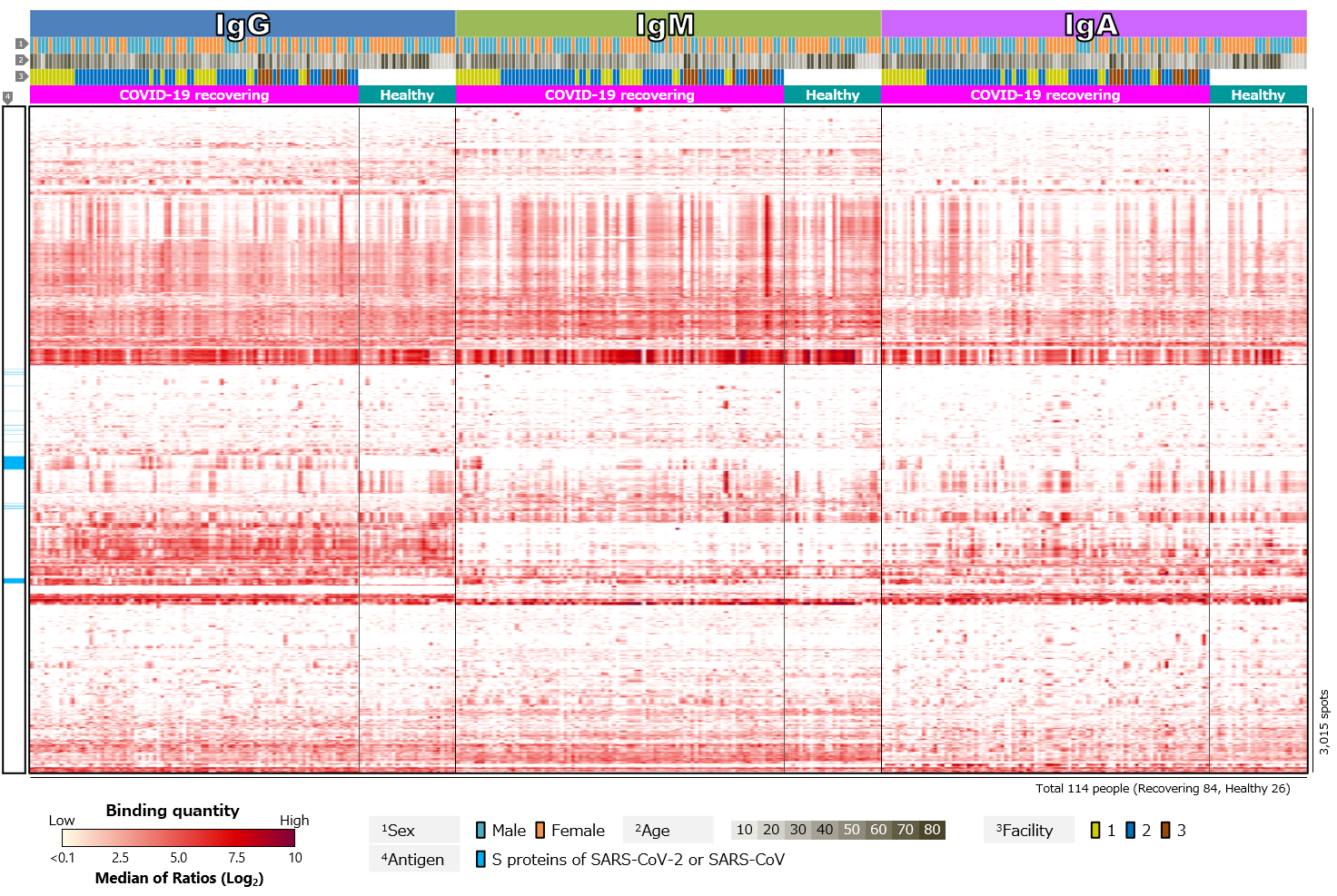
First, we compared blood antibody profiles against SARS-CoV-2 antigens in 84 COVID-19 recovered individuals and 26 healthy individuals (Figure 2). Antibodies against SARS-CoV-2 antigens were detected in most cases of COVID-19 recovered individuals, but not in the healthy individuals (Figure 2a). This indicates that the infection with SARS-CoV-2 induced antibodies against the virus. Focusing on the class of antibodies, IgG was positive in the largest number of cases. There was diversity in intensity of reactions and pattern of reacting antigens among the cases, with some cases showing little or no reactions and others having antibodies to a wide range of antigens. There was also a tendency for “male” and “older” specimens to respond more strongly. On the other hand, positive rates for IgM tended to be low. According to textbooks, IgM is first to react in early stages of infection, and then class switches to IgG and IgA. Thus, IgG and IgA appear later than IgM, and IgM disappears after a short period of time. Blood samples are taken from volunteers after a certain period of time has elapsed since infection. The overall low IgM positivity rate may be due to. However, number of days elapsed after infection varies from case to case, so it is assumed that those with a strong IgM response have had a relatively short period of time, while those with a weak response have had a long period of time since infection. Focusing on IgA, individual differences were observed, especially in antibodies against RBD. IgG against RBD was found in almost all cases, while IgA against RBD was found in a limited number of cases. IgA also tended to respond less to RBD than to non-RBD region of S1 subunit (N-terminal region) or to S2 subunit. In addition, there was a tendency for “older” individuals to respond more strongly (Figure 2b).
Click the figure to enlarge
Figure 2. Blood antibody profile against SARS-CoV-2 antigens of individuals recovering from COVID-19.
■ Analysis numerical data(Click the button to download)
| Title | File format |
| Blood antibody profile against SARS-CoV-2 antigens of individuals recovering from COVID-19. |
CSV |
| Excel | |
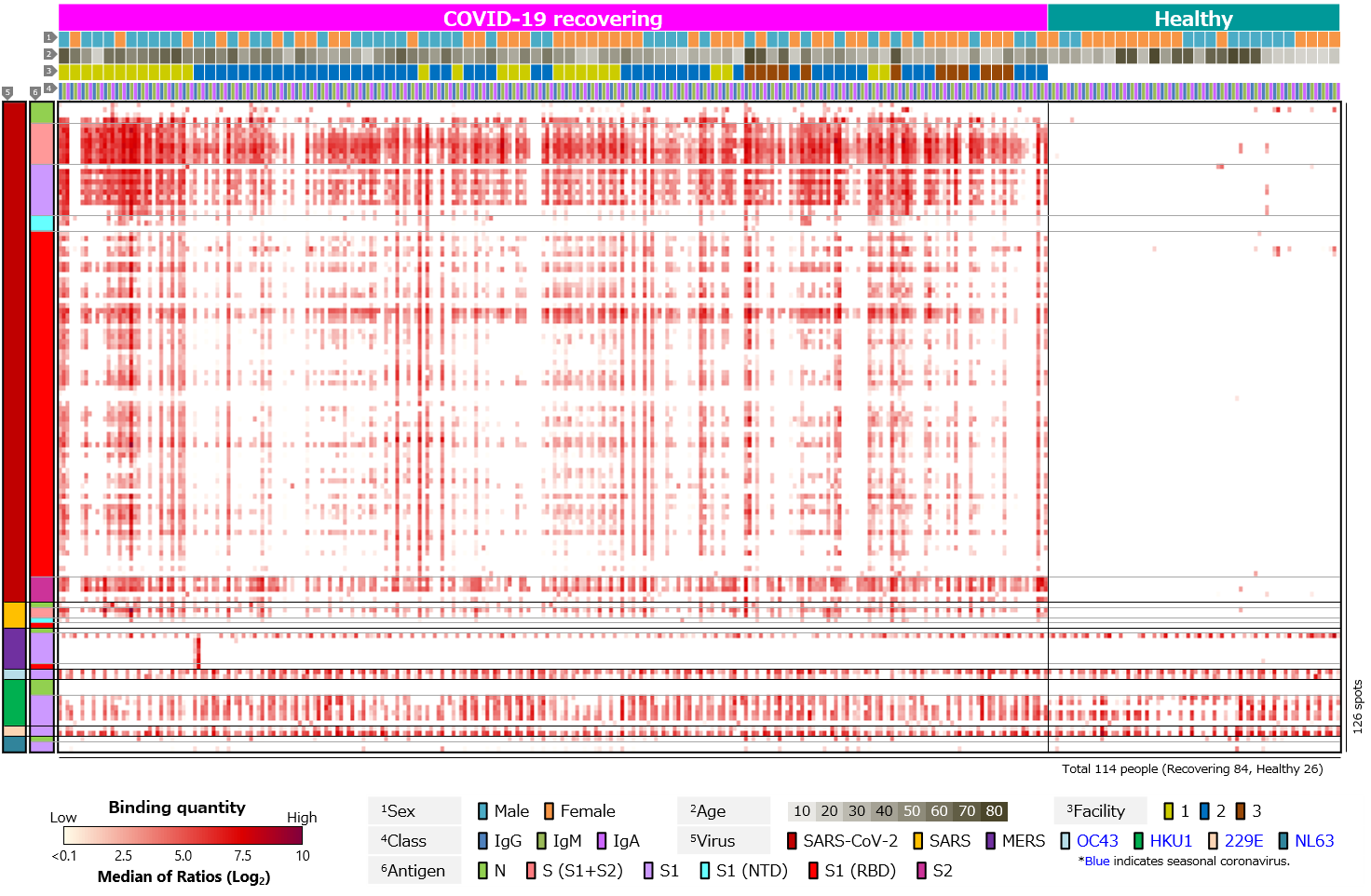
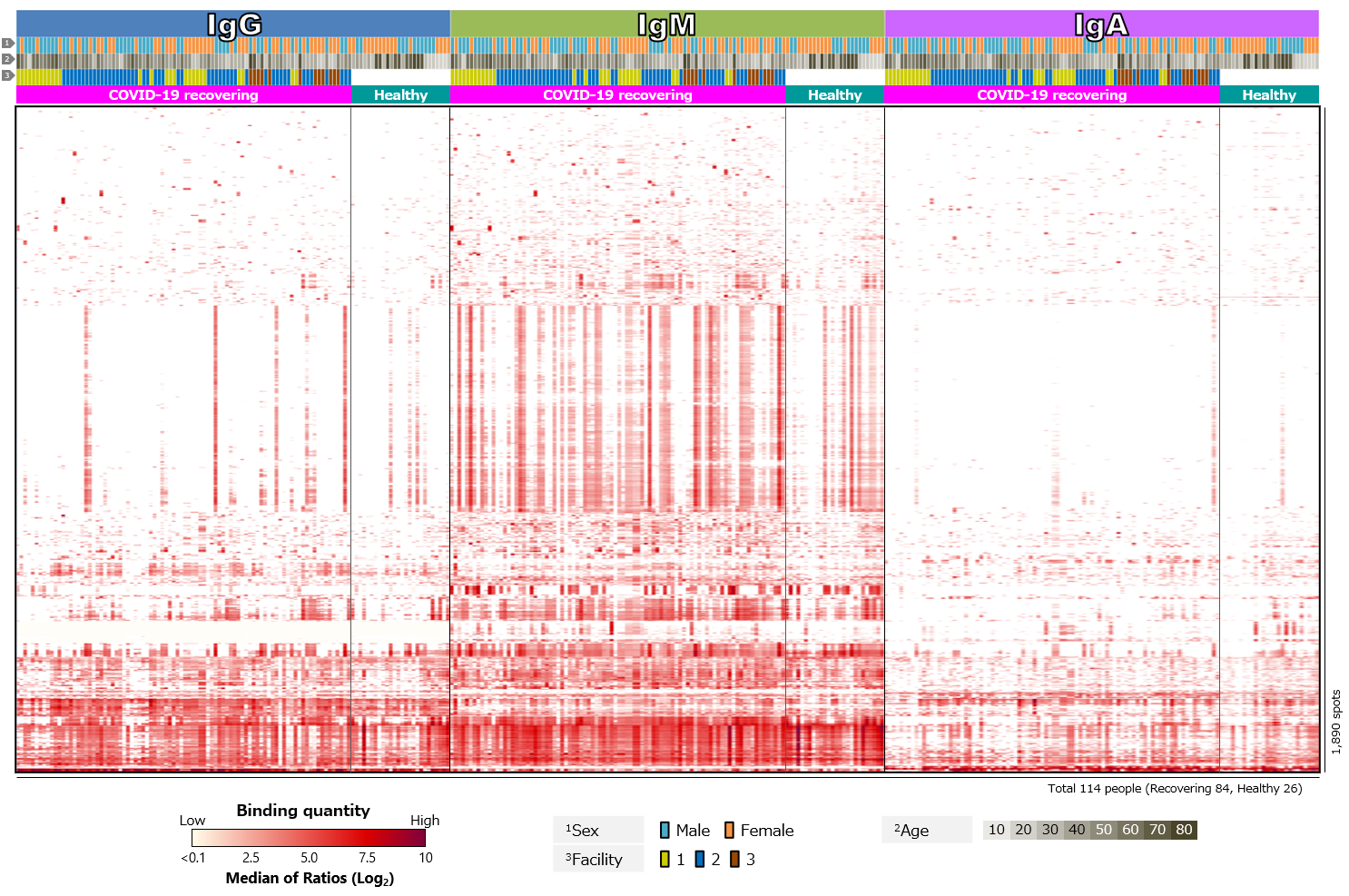
Second, blood antibody profiles against antigens of various human infectious coronaviruses, which are pathogens of severe pneumonia such as SARS and MERS, and seasonal colds, were compared among recovering and healthy individuals (Figure 3). Antibodies against common human coronaviruses were detected in healthy individuals, with no difference between those with COVID-19 recovered individuals (Figure 3a). However, few IgM responses to common coronaviruses have been detected (Figure 3b). We believe this is due to older infection date of common coronaviruses in all cases and complete switch in class from IgM to IgG or IgA.
Click the figure to enlarge
Figure 3. Blood antibody profile against coronavirus-related antigens of individuals recovering from COVID-19.
■ Analysis numerical data(Click the button to download)
| Title | File format |
| Blood antibody profile against coronavirus-related antigens of individuals recovering from COVID-19. |
CSV |
| Excel |
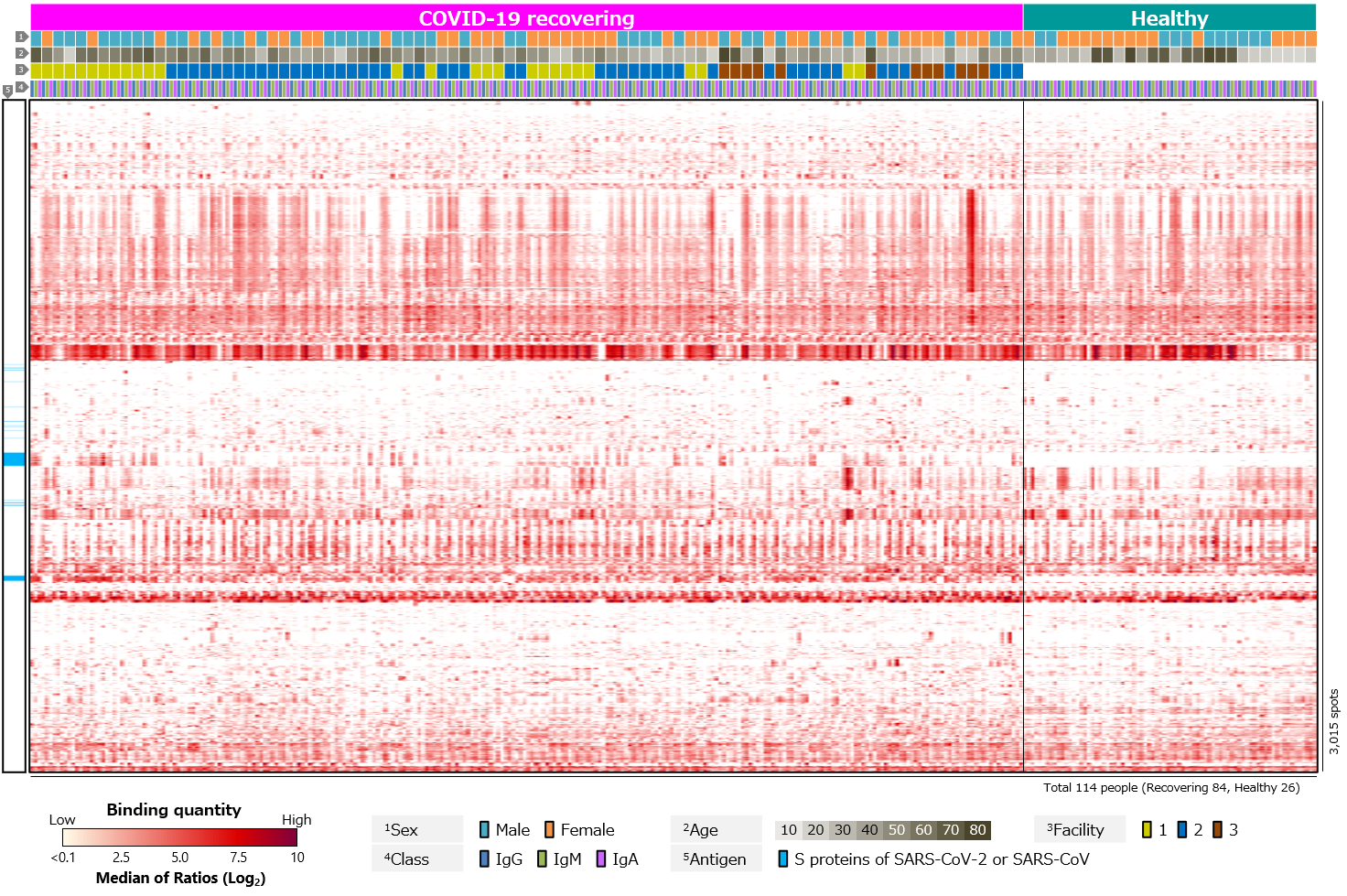
Third, blood antibody profiles against exogenous antigens, including various pathogenic microorganisms, commensal bacteria, and allergens, were compared (Figure 4). The results showed clear differences between recovering and healthy individuals for SARS-CoV-2 antigens. On the other hand, there were no clear differences for other antigens.
Click the figure to enlarge
Figure 4. Blood antibody profile against exogenous antigens of individuals recovering from COVID-19.
■ Analysis numerical data(Click the button to download)
| Title | File format |
| Blood antibody profile against exogenous antigens of individuals recovering from COVID-19(Excerpt only positive spots with at least one case) |
CSV |
| Excel | |
| Blood antibody profile against exogenous antigens of individuals recovering from COVID-19(All data) |
CSV |
| Excel |
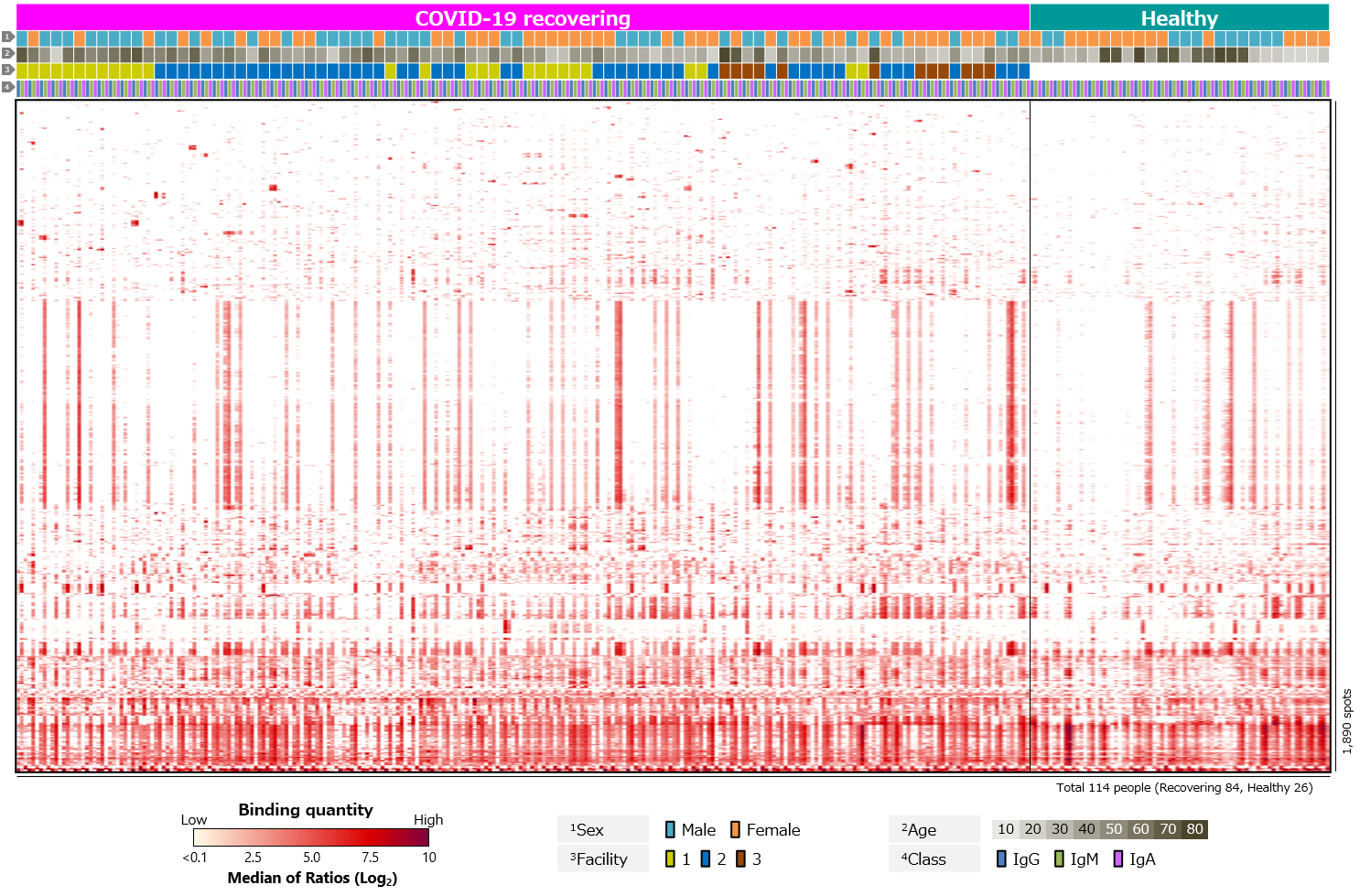
Finally, we analyzed blood antibody profiles against human proteins, i.e., autoantigens (Figure 5). While antibodies against SARS-CoV-2 act to protect against infection, there have been a number of reports of autoantibodies detected in COVID-19 patients. COVID-19 has been found to be more severe in elderly and those with underlying diseases such as diabetes. In addition, some people who have been infected with COVID-19 suffer from a chronic sequela called long COVID. Autoantibodies have been suggested as a cause or risk factor (Supplementary info. 3). It is believed that autoantibodies produced by some infection prior to infection with SARS-CoV-2 mistakenly attack own immune cells. However, the detailed mechanism is not yet clear. Analysis of blood antibody profiles against autoantigens in COVID-19 recovering individuals revealed that several antigens were positive in almost all cases. In addition, some antigens were detected specifically in each case, and some antigens were positive in multiple cases. There were also antigens with differential positivity rates between recovering and healthy individuals.
Click the figure to enlarge
Figure 5. Blood antibody profile against autoantigens of individuals recovering from COVID-19.
■ Analysis numerical data(Click the button to download)
| Title | File format |
| Blood antibody profile against autoantigens of individuals recovering from COVID-19(Excerpt only positive spots with at least one case) |
CSV |
| Excel | |
| Blood antibody profile against autoantigens of individuals recovering from COVID-19(All data) |
CSV |
| Excel |
Conclusion
Using the “protein microarray system” originally developed by the Fukushima project, we analyzed antibodies in blood of COVID-19 recovering individuals. Through this system, we were able to identify individual differences in the profiles of antibodies against exogenous antigens and autoantigens as well as antibodies against SARS-CoV-2. The unique feature of this system is that it can capture state of liquid immunity to a wide variety of antigens, not just the antigens of interest, all in one glass slide. The world is now receiving mRNA vaccination against SARS-CoV-2. Using the protein microarray, state of liquid immunity to not only the S protein of wild strain, which is the target of the vaccine, but also to S proteins of variants, and other viral antigens, microbial antigens, allergens, and autoantigens can be analyzed once and comprehensively. Comparative analysis of differences before and after vaccination, differences depending on whether or not an adverse reaction occurred after vaccination, and differences between vaccinated and naturally infected individuals would lead to the accumulation of useful and objective data for evaluating the effectiveness of vaccines. The system can also be applied to infectious diseases other than COVID-19. We are confident that our protein microarray system can be applied not only to analysis of influenza, which is a recurring epidemic worldwide every year, but also to analysis of new infectious diseases that may occur in the future.
Supplementary information
1) Development and approval status of neutralizing antibody drugs against SARS-CoV-2 In the U.S., antibody cocktail therapy combining two neutralizing monoclonal antibodies (casirivimab and imdevimab) developed by Regeneron, antibody combination therapy (bamlanivimab and etesevimab) developed by Eli Lilly, and sotrovimab co-developed by GlaxoSmithKline and Vir Biotechnology are approved as treatment for mild COVID-19 patients at high risk severe disease. In addition, AstraZeneca has applied for an emergency use authorization for an antibody cocktail (cilgavimab and tixagevimab). Antibody cocktail therapy with casirivimab/imdevimab and sotrovimab have been approved as fast track in Japan. The European Medicines Agency is expected to approve Regkirona, developed by Celltrion. Boehringer Ingelheim has developed BI 767551, which can be delivered directly to lungs via inhalation and has advanced to the clinical trial stage.
2) Classes of human antibodies There are five classes of human antibodies: IgG, IgM, IgA, IgD, and IgE. In the Fukushima project, research has focused on IgG, IgM, and IgA, which are deeply involved in mucosal membrane, a pathway through which pathogens such as viruses and bacteria enter human body, and in immune response after invasion. IgG is the most abundant antibody in blood and binds to pathogens such as viruses and bacteria that have invaded human body and inhibits their action. IgM forms a pentameric structure and is the first antibody produced when infected with a virus or bacteria. IgA exists as a monomer in blood and as a dimer on mucosal surface such as respiratory tract and intestinal tract and in in colostrum, and plays a role in preventing pathogens such as viruses and bacteria from entering a body. Because IgM and IgA form multimers, they are expected to bind to pathogens more effectively than IgG, which is a monomer. In a report published in Nature in June of this year, When the variable region of an IgG antibody with neutralizing activity against SARS-CoV-2 was combined with a constant region of IgM or IgA to produce a hybrid antibody, the efficiency of inhibiting viral infection wad increased compared to the original IgG antibody and an IgM antibody that acquired neutralizing ability against variants was obtained. This report further demonstrates efficacy of intranasal administration of IgM antibodies in animal study using mice.
3) Involvement of autoantibodies in severe cases of COVID-19 An international research team from Rockefeller University and others found that antibodies against type I interferon (type I IFN) at a very high frequency in COVID-19 deaths and in the most severe cases in elderly*1. In the report, autoantibodies against type I IFN were not found in mild or asymptomatic cases, while they were found in about 10% of severe cases, and more prevalent in older patients. Meanwhile, Yale and other research team has detected autoantibodies against extracellular and secreted proteins (exoproteome) in mild or asymptomatic COVID-19 patients and health care workers. In the report, they found increased autoantibody reactivity in COVID-19 patients compared to non-infected individuals, especially the prevalence of autoantibodies to immunomodulatory proteins*2. They also reported an increase in disease severity when antibodies against those target proteins were administered to a mouse model of SARS-CoV-2 infection. This means that autoantibodies play a role in enhancing the disease.
*1 P. Bastard et al., Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020).
*2 E. Y. Wang et al., Diverse functional autoantibodies in patients with COVID-19. Nature 595, 283-288 (2021).
■ Document(Click the button to download)
| Title | File format |
| Antibody profiling in blood of individuals recovering from COVID-19 |